Back to the Classics: Cell Sorting
How do cells orient themselves into organised structures while a tissue grows? This explorable describes the very first Cellular Potts Model (CPM): the cell sorting model of François Graner and James Glazier. After a bit of history, the interactive model will illustrate how cells can sort themselves into ordered spatial patterns—a key ability for processes like embryogenesis and tissue development.
A Brief History of the CPM
The first version of a CPM was developed in the early nineties by François Graner and James Glazier (Graner and Glazier, 1992). At the time, developmental biologists had found that when two different types of embryonic cells were mixed, they would sort into large, homogeneous same-cell patches—spontaneously! Experimental studies suggested that this sorting process arose from differences in adhesion between the cell types. To understand how this might work, Graner and Glazier wanted to model the phenomenon and see if they could indeed reproduce sorting based on such "differential adhesion" alone (in other words: they wanted to test the so-called differential adhesion hypothesis).
Inspiring Magnets
Their model was an extension of the existing Potts model, originally developed by physicists to simulate magnets. The Potts model simulates the interaction between so-called "spins" on a crystalline lattice.
The idea is simple: we have a grid with a bunch of pixels, representing points on the crystal lattice. These points all have a property called their spin, which can be either "down" (0) or "up" (1). They then interact as follows:
- we assign an energetic penalty to each pair of neighbor pixels with differing spin;
- pixels continuously try to change (or "flip") their spin, but...
- they are more likely to succeed if this is energetically favourable (i.e. if the flip decreases the number of opposite-spin neighbors).
For more details on the model, see this tutorial; the model below is essentially the model from that tutorial with Adhesion only. For now though, let's just look at the result (with red and gray representing the "up" and "down" spins):
While the Potts model does not much look like a cell, Graner and Glazier noticed that it does have an interesting property: the spins on the lattice automatically sort into large patches of same-spin sites. And even the mechanism somewhat resembled their differential adhesion hypothesis: the energetic cost for having adjacent pixels with opposite spin could be interpreted as a "contact energy" or "adhesion". Could they apply a similar principle to the cell sorting question?
The Cellular Potts Model
A problem with the Potts model is that pixels can only have one of two states, since magnetic spins can only be "up" or "down". While you could interpret this as "cell" or "empty background" instead, Graner and Glazier needed to model many different cells for their differential adhesion simulation—not just a single cell on an empty background. Their model replaced the binary spin property with a "cell identity", a number indicating the cell to which a grid point belonged. (The trick here is that in contrast to the spin, the cell identity number could be any integer \(\geq\)0 rather than only 0 or 1—allowing the co-existence of multiple cells on the same grid).
A second problem is that the patches formed in the Potts model can be of any size and shape, unlike cells. Graner and Glazier therefore extended the energy equations so that "cells" would roughly keep the same size (number of grid points), by assigning energetic penalties for any deviations from this "target volume" (see this tutorial for details; Graner and Glazier, 1992, Glazier et al, 2007).
They then used this to model their differential adhesion hypothesis. They did this in a similar way as the original Potts model described earlier, but now:
- Instead of the binary "spin" property (0 or 1), pixels can now have a value \(v\) in the range of 0-\(n\), where \(n\) is the number of cells
- Each cell tries to maintain its size as described above
- Each pixel also has a second property \(k\), describing the kind of "cell" it belongs to: the empty background (0) or one of the two self-sorting cell types 1 and 2
- Like with the spins, we assign a penalty \(J\) to each pair of neighbor pixels that do not belong to the same cell (i.e. have different \(v\)). But now the size of this penalty depends on the kind \(k\) of the contacting pixels: we have \(J_{\text{bg,kind 1}}\), \(J_{\text{bg,kind 2}}\), \(J_{\text{kind 1, kind 1}}\), \(J_{\text{kind 2, kind 2}}\), \(J_{\text{kind 1, kind 2}}\), and
- To encode the "differential adhesion":
$$J_{\text{kind 1, kind 1}}, J_{\text{kind 2, kind 2}} \lt J_{\text{kind 1, kind 2}}$$(i.e., assign a larger penalty to cell contacts if the cells belong to different types)
The result was pretty magical:
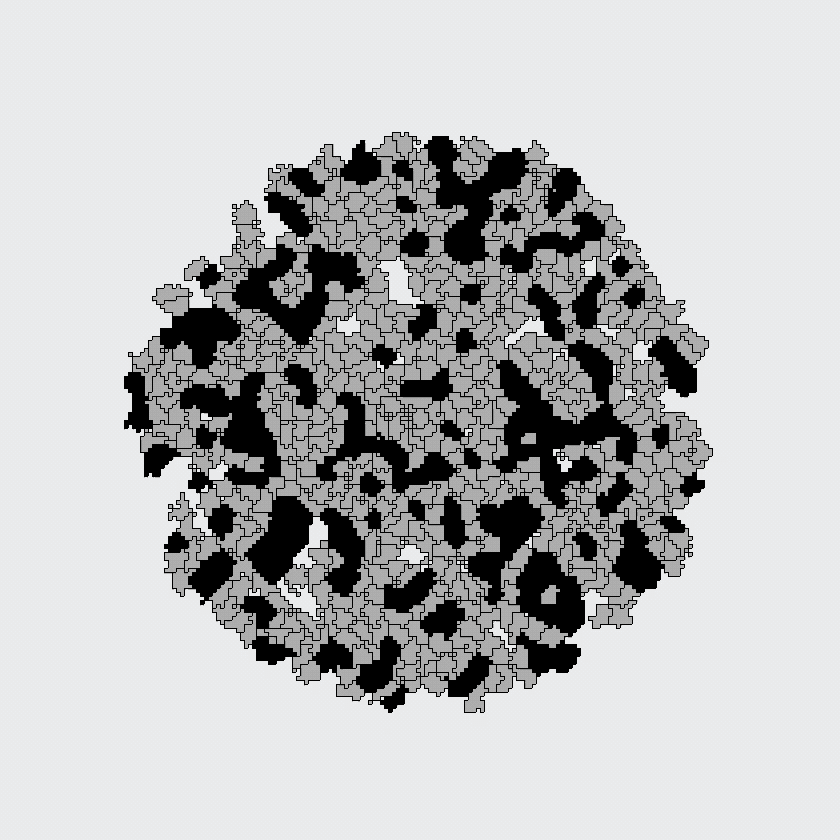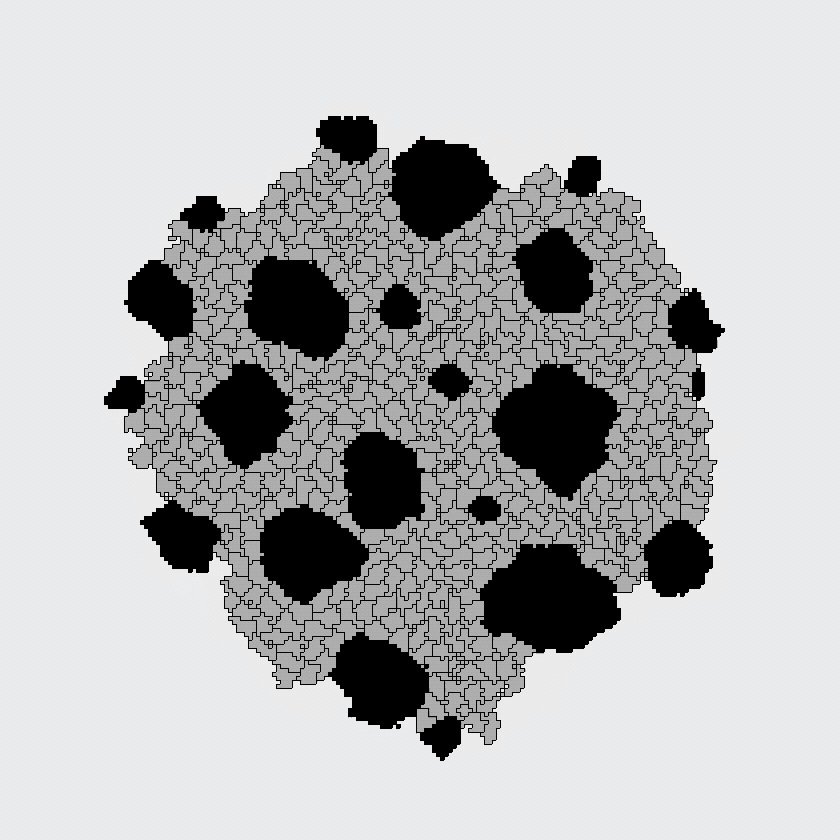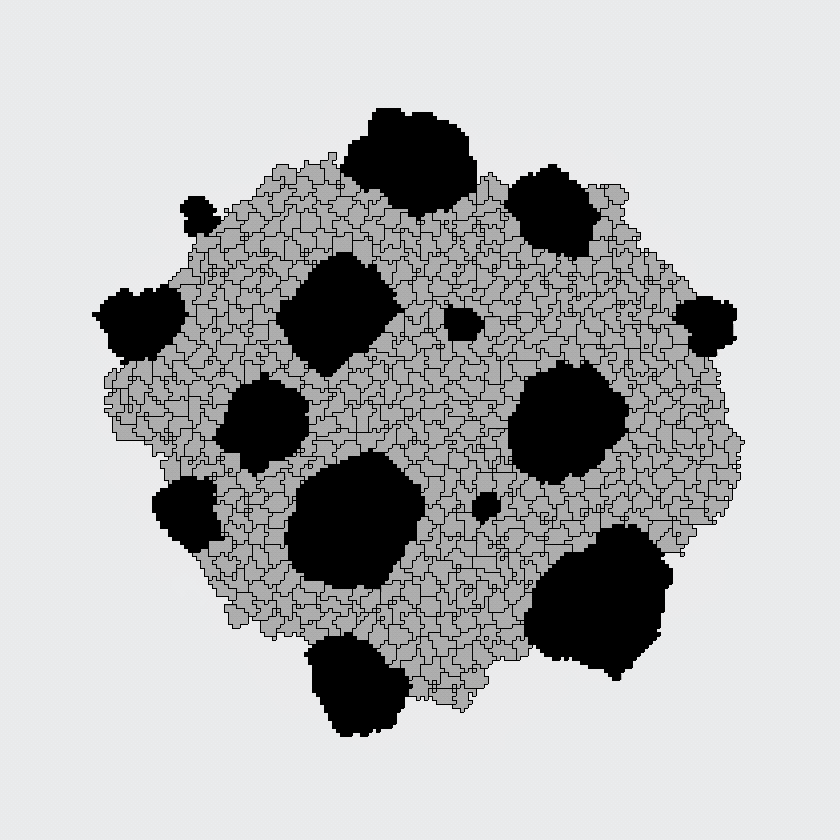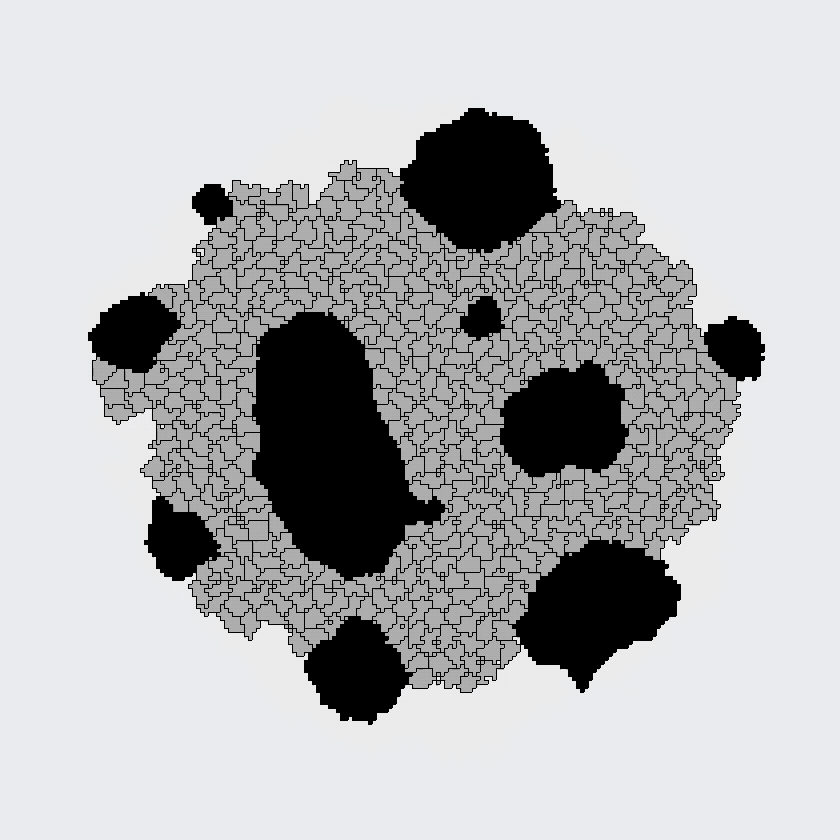
Here, the black and gray cells start out randomly mixed, but automatically sort themselves into large, same-cell patches. This simulation supported the differential adhesion hypothesis: apparently, cells could find each other based on adhesion alone, because this model contained no other mechanism for cells to find each other.
Let's now explore the cell sorting behaviour in more detail, using an interactive model.
Try It Yourself
Below, you'll find Graner and Glaziers model of cells sorting through differential adhesion. You can adjust the different adhesion parameters using the sliders below.
| Jbg,gray | 0 |
|
30 |
| Jbg,red | 0 |
|
30 |
| Jgray,gray | 0 |
|
30 |
| Jred,red | 0 |
|
30 |
| Jgray,red | 0 |
|
30 |
Suggestions:
- Click play (►) and let the simulation run for a while. You should see that most of the cells on the outside of the "blob" are red; this is because the gray cells have a higher contact penalty with the background than the red cells do, so the gray cells prefer to "hide" behind the red ones.
- Set Jbg,gray to 6 and Jbg,red to 12. What happens and why?
- Now increase Jbg,red further, to 30. You'll see that the gray cells on the outside stretch out; preventing the red cells from contacting the background becomes more important than preventing the gray cells from falling apart.
- Set both Jbg,red and Jbg,gray to 8, and increase Jgray,red to 12. You should see that cells start sorting. Once they have, you can click on "blender" to see them do it again (just to make sure it wasn't a fluke...)
- What happens when you set Jgray,red to its maximum value?
- Now set both Jbg,red and Jbg,gray to 8, Jred,red and Jgray,gray to 14, and Jred,gray to 6. What do you see?
Summary
The CPM was developed in the nineties as an extension of an existing model of magnetism, in which dynamics arise from an energetic optimization process. The model of Graner and Glazier considered only energies associated with cell-cell adhesion and with cell size, and was designed to answer a specific question: can cells of different types sort themselves in space based on "differential adhesion" alone? Indeed, spatial patterning arose spontaneously in the Graner-Glazier model, showing that differential adhesion was sufficient to explain the cell sorting process.
This was the first "cellular" Potts model, or CPM. After Graner and Glazier showed how the central energy equation of the original Potts model could be extended to incorporate processes relevant to cells, others quickly followed with their own variations to the cellular Potts model. In particular, Paulien Hogeweg developed important extensions to generalize the CPM—which is why it is also referred to as the Glazier-Graner-Hogeweg (GGH) model (Glazier et al, 2007).